

Alternative manual methods are available and are described in great detail by Edman (1950), and reviewed by Niall (1973), Allen (1981), and Schlesinger (1988) (see Further Reading). The PTH amino acids are then identified most commonly by liquid chromatography (LC). The TFA-treated thiazolinone amino acid is extracted with 0.15 ml of benzene and, following drying, is converted to its PTH isomer in 0.2 ml of 1 mol l −1 HCl at 80☌ for 10 min.

Cleavage products are dried under a stream of nitrogen for 30 s. Cleavage is accomplished with 75 μl of TFA under nitrogen at 54☌ for 5 min. Following coupling, a single extraction with 0.1 ml of benzene is performed and the organic phase is discarded. In the manual mode, coupling of the peptide is performed under nitrogen in a stoppered 5-ml conical centrifuge tube for 30 min at 54☌ in 100 μl of 0.4 mol l −1 dimethylallylamine (DMAA) buffer in propanol–water (60:40 v/v), previously adjusted to pH 9.5 with TFA. The coupling medium is a mixture of aqueous and organic solvents to enhance the solubility of both the peptide and the PITC reagent. Schlesinger, in Encyclopedia of Analytical Science (Second Edition), 2005 Edman Chemistry and the Manual MethodĮdman degradation is a three-step procedure consisting of the coupling of phenylisothiocyanate (PITC) to the α-amino group of a peptide or protein, cleaving the amino-terminal amino acid (via cyclization in strong per-fluorinated acid, typically trifluoroacetic acid (TFA), to a 2-anilino-5-thiazolinone), and converting the resulting thiazolinone in aqueous acid to its more stable isomer, the phenylthiohydantion (PTH) amino acid.Ĭoupling takes place most rapidly and completely under alkaline conditions at a pH above the p K of the amino group (pH=7.8), at which it is uncharged, and at pH below 10, above which PITC is hydrolyzed. Presented herein is the detailed step-by-step protocol for ATOMS.ĭ.H. We successfully used ATOMS to identify nearly 100 cleavage sites in the ECM proteins laminin and fibronectin. ATOMS employs isotopic labeling and quantitative tandem mass spectrometry to identify cleavage sites in a fast and accurate manner. We recently developed Amino-Terminal Oriented Mass spectrometry of Substrates (ATOMS) to overcome these limitations as a complement for N-terminal sequencing. N-terminal microsequencing of proteolytic fragments is the usual method employed, but it suffers from poor resolution of sodium dodecylsulfate-polyacrylamide gel electrophoresis gels and is inefficient at identifying multiple cleavages, requiring preparation of numerous gels or membrane slices for analysis.

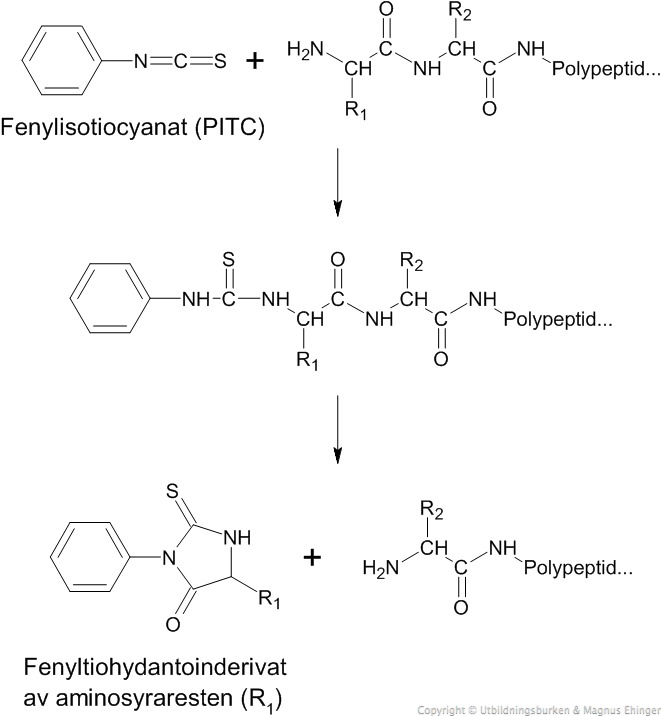
However, the identification of cleavage sites in complex high molecular proteins such as those composing the ECM is not trivial. The exact proteolytic cleavage sites need to be identified to fully understand the functions of the cleavage fragments and biological roles of proteases in vivo. For example, extracellular matrix (ECM) protein processing often produces multiple proteolytic fragments with the generation of cryptic binding sites and neoproteins by ECM protein processing being well documented. Proteolytic processing can change the activity of bioactive proteins and also reveal cryptic binding sites and generate proteins with new functions (neoproteins) not found in the parent molecule. We describe a new method for the high-throughput determination of N-terminal sequences of multiple protein fragments in solution. However, for accurate data analysis and amino acid assignments, Edman sequencing proceeds on samples of single proteins only and so lacks high-throughput capabilities. Overall, in Methods in Enzymology, 2011 AbstractĮdman degradation is a long-established technique for N-terminal sequencing of proteins and cleavage fragments.


 0 kommentar(er)
0 kommentar(er)
